836 views
We just submitted a pre-print 'A novel variant of interest (VOI) of SARS-CoV-2 with multiple spike mutations detected through travel surveillance in Africa.'
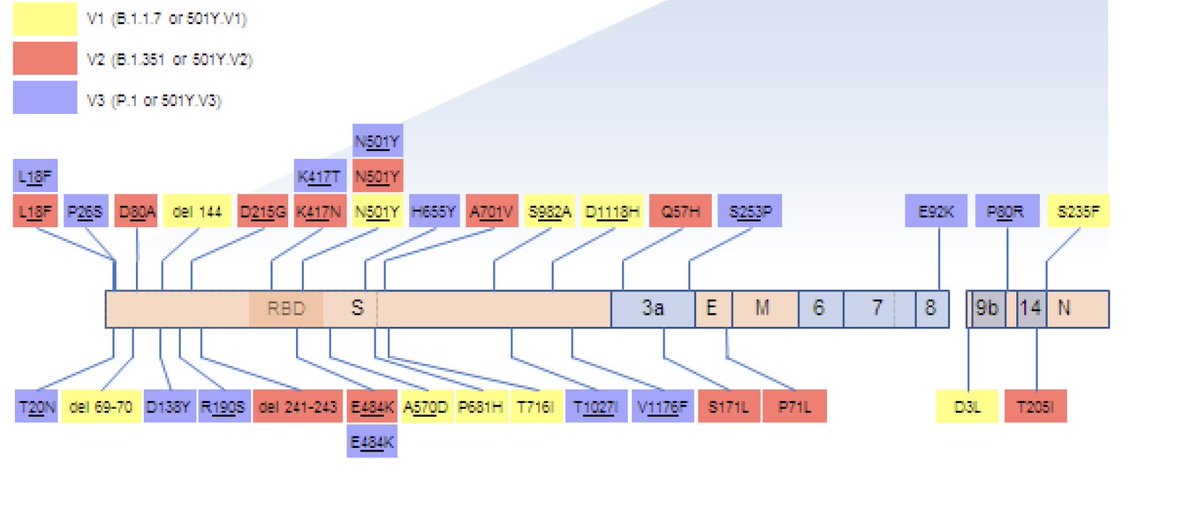
This VOI has 31 amino acids mutations. In Spike has 11 mutations and three deletions in the N-terminal domain
This VOI has 31 amino acids mutations. In Spike has 11 mutations and three deletions in the N-terminal domain
It has some key mutations, including the E484K, R346K and P681H. The R346K is the associated with resistance to class 3 RBD NAbs recently described by @jbloom_lab
There are also 5 substitutions and 3 deletions in the NTD antigenic supersite (Y144Δ, R246M, SYL247-249Δ and W258L)
There are also 5 substitutions and 3 deletions in the NTD antigenic supersite (Y144Δ, R246M, SYL247-249Δ and W258L)
It the most diverse A lineage sequencers ever described. It also worry us as it was found in three travelers from Tanzania in Angola. There is almost no data from COVID-19 in Tanzania. 
When compared with other variants of concern and variants of interest, this is the most divergent one. 
Manuscript has been deposited at medRxiv and until online is freely accessible at: krisp.org.za/publications.p…
Sequences are available in GISAID with accessions: EPI_ISL_1347940, EPI_ISL_1347941, EPI_ISL_1347942
Sequences are available in GISAID with accessions: EPI_ISL_1347940, EPI_ISL_1347941, EPI_ISL_1347942
We decided to report this as a new VOI given the constellation of mutations with known or suspected biological significance, specifically resistance to neutralizing antibodies and potentially increased transmissibility.
Whilst we have only detected three cases with this new VOI, this warrants urgent investigation as the source country, Tanzania, has a largely undocumented epidemic and few public health measures in place to prevent spread within and out of the country.
Thanks @AfricaCDC, Minister of Health of Angola, @rjlessells @houzhou @Mittenavoig and many other colleagues from Angola and Africa to help on the analysis of the data.
• • •
Missing some Tweet in this thread? You can try to
force a refresh